Steps of DNA Replication
Wednesday, April 1, 2009
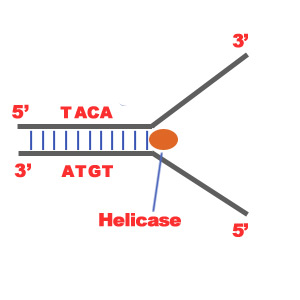
1)The first major step for the DNA Replication to take place is the breaking of hydrogen bonds between bases of the two antiparallel strands. The unwounding of the two strands is the starting point. The splitting happens in places of the chains which are rich in A-T. That is because there are only two bonds between Adenine and Thymine (there are three hydrogen bonds between Cytosine and Guanine). Helicase is the enzyme that splits the two strands. The initiation point where the splitting starts is called "origin of replication".The structure that is created is known as "Replication Fork".

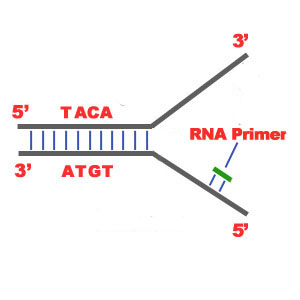
2) One of the most important steps of DNA Replication is the binding of RNA Primase in the the initiation point of the 3'-5' parent chain. RNA Primase can attract RNA nucleotides which bind to the DNA nucleotides of the 3'-5' strand due to the hydrogen bonds between the bases. RNA nucleotides are the primers (starters) for the binding of DNA nucleotides.






0 comments:
Post a Comment